Machine Learning Primer for Clinicians–Part 10
Alexander Scarlat, MD is a physician and data scientist, board-certified in anesthesiology with a degree in computer sciences. He has a keen interest in machine learning applications in healthcare. He welcomes feedback on this series at drscarlat@gmail.com.

Previous articles:
- An Introduction to Machine Learning
- Supervised Learning
- Unsupervised Learning
- How to Properly Feed Data to a ML Model
- How Does a Machine Actually Learn?
- Artificial Neural Networks Exposed
- Controlling the Machine Learning Process
- Predict Hospital Mortality
- Predict Length of Stay
While previous articles have described supervised ML models of regression and classification, in this article, we’re going to detect anomalies in antibiotic resistance patterns using an unsupervised ML model.
By definition, anomalies are rare, unpredictable events, so we usually don’t have labeled samples of anomalies to train a supervised ML model. Even if we had labeled samples of anomalies, a supervised model will not be able to identify a new anomaly, one it has never seen during training. The true magic of unsupervised learning is the ML model capability to identify an anomaly never seen.
The Python code and the dataset used for this article are available here.
Data
The data for this article is based on a subset of MIMIC3 (Multiparameter Intelligent Monitoring in Intensive Care), a de-identified, freely available ICU database, Using SQL as detailed in my book , a dataset of 25,448 antibiograms was extracted. The initial dataset includes 140 unique microorganisms and their resistance / sensitivity to 29 antibiotics arranged in 25,400 rows:
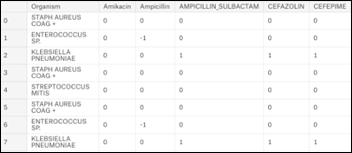
- +1 = organism is sensitive to antibiotic
- 0 = information is not available
- -1 = organism is resistant to antibiotic
Summarizing the above table by grouping on the organisms produces a general antibiogram:
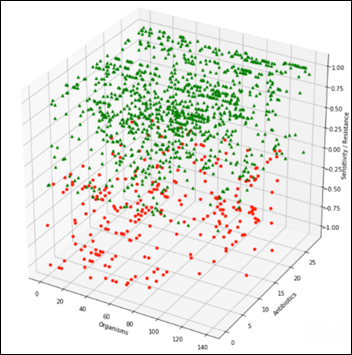
A view perpendicular to the organisms axis on the above chart becomes the projection of all the organisms and their relative sensitivity vs. resistance to all antibiotics:

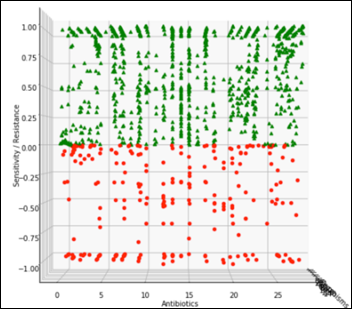
The view perpendicular to the antibiotics axis becomes the projection of all the antibiotics and the relevant susceptibility of all the organisms:
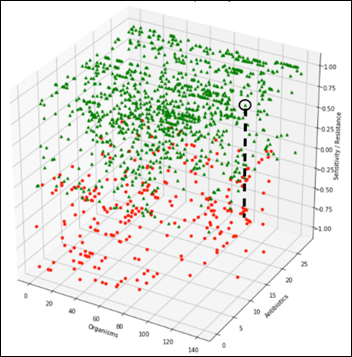
The challenge is to detect an anomaly that may manifest itself as a slight change of an organism susceptibility to one antibiotic, for example, along the black dashed line in the diagram below. During testing of the ML model, we’ll gradually modify one organism susceptibility to one antibiotic and test the model on F1 score, repeatedly, at different levels of susceptibility:
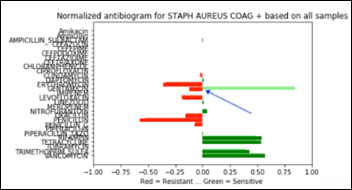
Let’s focus on the most frequent organism in the data set – Staph Aureus Coag Positive (Staph).
Staph has 6,925 samples out of 25,400 (27.2 percent of the whole dataset) and its average antibiogram in a projected 3D view:
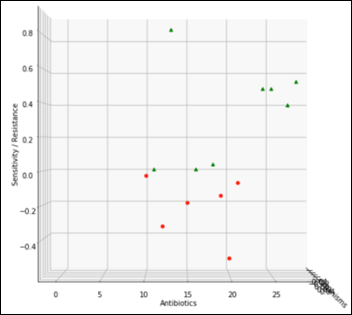
The above chart projected on 2D and rotated 90 degrees clockwise depicts the average antibiogram of Staph:
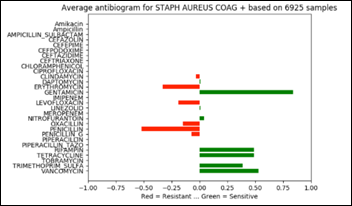
Note that in the above diagram, zero has an additional meaning besides “information not available.” Zero may be also the result of averaging a number of sensitive with the same number of resistant samples and thus and average of zero susceptibility.
Model
- Task: unsupervised anomaly detection in antibiograms, a binary decision: normal vs. anomaly
- Experience: 25,400 antibiograms defined as normal for model training purposes
- Performance: F1 score at various degrees of anomalies applied to the average antibiogram of one organism (Staph) and one antibiotic at a time
Local Outlier Factor (LOF) is an anomaly detection algorithm introduced in 2000, which finds outliers by comparing their location with respect to a given number of neighbors (k). LOF takes a local approach to detect outliers about their neighbors, whereas other global strategies might not be the best detection for datasets that fluctuate in density.
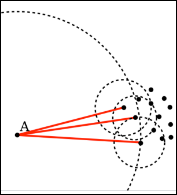
If k = 3, then the point A in the above diagram will be considered an outlier by LOF, as it is too far from its nearest three neighbors.
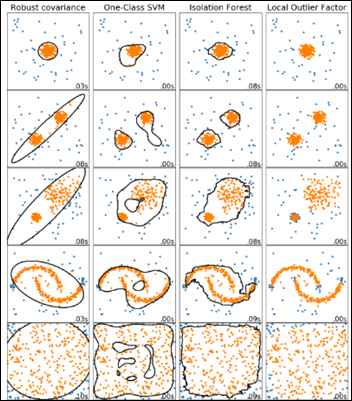
A comparison of four outlier detection algorithms from scikit-learn on various anomaly detection challenges: yellow dots are inliers and blue ones are outliers, with LOF in the rightmost column below:
The LOF model is initially trained with the original antibiograms dataset, which the model will memorize as normal.
After the model was trained, we gradually modify the Staph susceptibility to:
- Vancomycin only: from the original +0.5 to 0.4, 0.2, 0, -0.2,… -1.0
- Gentamicin only: from the original +0.8 to 0.4, 0.2, 0, -0.2,… -1.0
- Both antibiotics at the same time: 0.4, 0.2, 0, -0.2,… -1.0 sensitivity / resistance
Below is a Staph antibiogram with only 13 percent sensitive to gentamicin compared to the normal Staph antibiotic susceptibility at 83 percent for the same antibiotic:
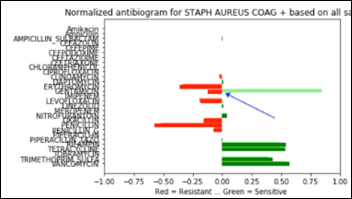
And the same comparison as above, but with a Staph population that is 13 percent resistant to gentamicin:
Performance
The last example, Staph susceptibility to gentamicin significantly shifting from (+0.83) to (-0.13) creates a confusion matrix with the following performance metrics:
- Accuracy: 87.3 percent
- Recall: 82.6 percent
- Precision: 91.3 percent
- F1 score: 0.867
At each antibiotic sensitivity / resistance level applied as above, the model performance is measured with F1 score (the harmonic mean of precision and recall detailed in previous articles). The model performance charted over a range of Staph susceptibilities to vancomycin, gentamicin, and both antibiotics:
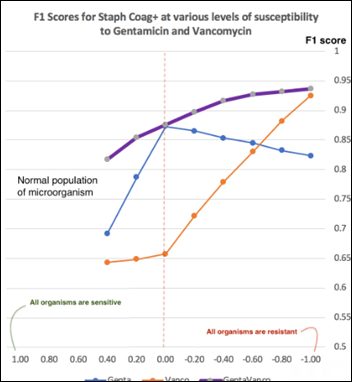
The more significant an anomaly of an antibiogram, the higher the F1 score of the model. An anomaly may manifest itself as a a single large change in the sensitivity of one organism to one antibiotic or as several small changes in the resistance to multiple antibiotics happening at the same time. A chart of LOF model performance over a range of anomalies can provide insights into the model capabilities at a specific F1 score. For example, at F1 = 0.75, Staph sensitivity to gentamicin declining from (+0.83) to (+0.2) will be flagged as an anomaly, but the same organism changing its vancomycin sensitivity from (+0.53) to (-0.2) will not be flagged as an anomaly.
There are no hard coded rules in the form of “if…then…” when using an unsupervised ML model. As there are no anomalies to use as labeled samples, there is a need to synthetically create outliers for testing the model performance by modifying samples features in what (we believe) may simulate an anomaly.
In all the testing scenarios performed with the LOF model, these synthetic anomalies have always been in one direction: from an existing level of sensitivity towards a more resistant organism, as this is the direction the bacteria are developing under the evolutionary pressures of antibiotics. A microorganism developing a new sensitivity towards an antibiotic is practically unheard of, as it dooms the bacteria to commit suicide when exposed to an antibiotic to which it was previously resistant.
Unsupervised anomaly detection is a promising area of development in AI, as these ML models have shown their uncanny, magic capabilities to sift through large datasets and decide, under their own volition, what’s normal and what should be considered an anomaly.
Next Article
Basics of Computer Vision


Seems the FTC ruling on non-competes hits start-ups hardest. All they've got are ideas and if staff leave with them…